Medical image segmentation is one of the most influential fields in modern healthcare AI, powering diagnostics, precision medicine, and real-time clinical decision support. Behind every robust model lies at least one medical segmentation dataset with carefully crafted labels that capture anatomical detail, pathology boundaries, and real clinical variation. The quality of these datasets directly shapes how AI systems perform in real hospitals and laboratories. For researchers, clinicians, and industry teams, understanding the strengths and limitations of each dataset is essential for building safe and reliable models.
Public biomedical datasets have grown rapidly over the last decade, supported by challenges organized by institutions like MICCAI, TCIA, and leading research hospitals. These initiatives help standardize evaluation protocols and improve reproducibility across studies. The Radiological Society of North America provides extensive educational and scientific imaging resources that further support this ecosystem.
At the same time, more specialized datasets have emerged for tasks such as tumor delineation, whole-organ labeling, vascular segmentation, and cell-level ground truth. Each provides unique insights for training and validating algorithms that must work under real clinical conditions.
Below is a deep, clinically grounded and technically accurate overview of the most important datasets used today for segmentation in radiology, digital pathology, microscopy, and multimodal medical imaging.
Why High Quality Segmentation Datasets Matter in Clinical AI
High performance segmentation models require more than diverse images. They need consistent ground truth masks created by radiologists, pathologists, biomedical engineers, and annotation specialists working under controlled quality workflows. These masks guide models to distinguish tissues that may differ by only a few grayscale values, such as subtle tumor boundaries or inflamed regions in soft tissue. When ground truth varies significantly between annotators, model error increases and clinical safety is compromised.
Medical image segmentation datasets also capture clinical domain shift, one of the biggest challenges in healthcare AI. Variations in scanner type, slice spacing, acquisition protocol, contrast timing, and patient anatomy all influence how models generalize. A dataset that includes multiple imaging vendors, field strengths, and patient demographics helps reduce brittle performance. By contrast, training exclusively on a single institution can create models that fail when deployed elsewhere.
High quality datasets provide standardized benchmarks, enabling teams to evaluate the maturity of their models against the broader scientific community. Competitions such as the Liver Tumor Segmentation Challenge or the Brain Tumor Segmentation Challenge have become essential in validating whether a model reaches acceptable accuracy for clinical decision support. To support biomedical research innovation, the National Institute of Biomedical Imaging and Bioengineering offers high quality scientific and technical resources.
What Defines a High Quality Medical Segmentation Dataset
A dataset suitable for clinical-grade AI must meet rigorous expectations on several axes. While each dataset has its own design, the strongest ones share key attributes that improve model robustness and safety.
Annotation Quality and Clinical Expertise
High quality datasets rely on labels created or validated by experienced clinicians who understand anatomical boundaries and pathological patterns. In organ segmentation, a few millimeters of deviation can change volumetric calculations and treatment planning margins. In tumor segmentation, high inter-observer agreement is essential for reproducibility. Datasets that document annotator experience, review protocols, and consensus methods provide stronger reliability.
Imaging Diversity and Technical Conditions
A meaningful dataset for medical imaging segmentation includes variability across scanners, vendors, field strengths, acquisition planes, and patient cases. This diversity helps reduce model overfitting to a narrow data distribution. Variability is especially critical in MRI, where imaging appearance differs widely between institutions and sequences. Datasets that include high-quality metadata, sequence names, voxel spacing, and modality parameters also make preprocessing more reproducible.
Clear Labels and Clinically Relevant Classes
A dataset is only as useful as its class definitions. Strong datasets provide segmentation masks that align with clinical tasks such as identifying liver lesions, mapping white matter changes, or delineating organs at risk for radiotherapy. Some datasets provide multi-label masks that allow models to learn hierarchical structures, which is particularly useful for whole-body or organ-system segmentation.
Documented Preprocessing and Benchmarking
Public datasets are most practical when accompanied by clear documentation of preprocessing requirements. Standardized pipelines, reference code, and train-test splits reduce variability across research studies. When datasets participate in challenges, they often include benchmark leaders, enabling teams to evaluate where their model stands relative to state-of-the-art approaches.
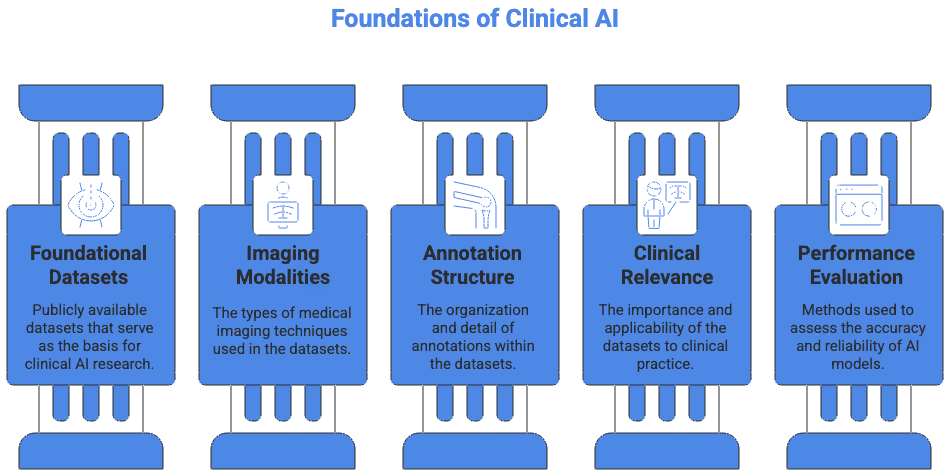
Foundational Datasets for Medical Imaging Segmentation
The following datasets represent some of the most widely used public datasets in clinical AI research. They cover a range of modalities including CT, MRI, ultrasound, and microscopy. Each provides unique segmentation challenges that help train robust models.
Brain Tumor Segmentation (BraTS)
The BraTS dataset is one of the most influential resources in modern medical imaging AI. Hosted as part of the MICCAI Brain Tumor Segmentation Challenge, it focuses on gliomas, which are among the most complex brain tumors to delineate. BraTS includes multi-parametric MRI sequences such as T1, T1-post contrast, T2, and FLAIR for each case, allowing models to leverage complementary information from different tissue contrasts.
BraTS uses consensus segmentations from experienced neuroradiologists, ensuring clinically meaningful boundaries. The dataset includes multiple tumor subregions such as enhancing tumor, edema, and necrotic core. This multi-label structure makes it ideal for training deep learning models that must support treatment monitoring or surgical planning. BraTS also provides strong benchmark leaderboards, enabling teams to compare performance over time and track improvements.
Liver Tumor Segmentation (LiTS)
The LiTS challenge dataset has become a standard reference for liver and lesion segmentation in CT imaging. Liver tumors show significant variability across patients, ranging from small hypodense metastases to large hepatocellular carcinomas. CT scans used in LiTS capture this diversity and help researchers build models that operate under realistic clinical variation.
Annotations in LiTS include both liver boundaries and individual tumor masks. This dual-label structure allows researchers to train models capable of both organ localization and lesion detection. Many studies use LiTS as a baseline for liver-related segmentation tasks due to its clean formatting and strong community benchmarks. It is highly valuable for developing models in oncology, surgical planning, and volumetric assessment.
ACDC Cardiac MRI Segmentation
The ACDC dataset focuses on short-axis cardiac MRI, including manually segmented contours for the left ventricle, right ventricle, and myocardium. These structures are essential for measuring cardiac function, ejection fraction, and wall thickness. ACDC helps researchers build models capable of segmenting dynamic sequences across multiple timepoints, reflecting real clinical motion and anatomical change.
ACDC includes cases with different cardiac conditions, making it a strong resource for generalizing across patient demographics. Variability across scanners and acquisition protocols further supports robust model development. Because cardiac imaging requires precise pixel-level accuracy, the high quality of the labels in this dataset makes it especially valuable for clinical applications. Several related resources can also be found through PhysioNet, a trusted MIT-managed platform hosting clinical imaging datasets.
Pancreas CT Dataset
The pancreas is one of the most difficult abdominal organs to segment due to its variable shape, surrounding tissues, and low contrast in CT imaging. The public pancreas CT dataset includes manually segmented contours created by radiology experts, capturing subtle anatomical boundaries. Models trained on this dataset help support downstream tasks such as lesion detection and postoperative evaluation.
Compared to liver or kidney datasets, the pancreas dataset poses a challenging benchmark for algorithm developers. Its relatively small sample size emphasizes the need for data augmentation, transfer learning, and careful validation. Teams working on abdominal segmentation often use this dataset to evaluate whether their models can handle low contrast organs.
Spleen CT Dataset
The spleen CT dataset is frequently used in multi-organ segmentation research. Spleen segmentation is clinically relevant for assessing trauma, splenic injury, or enlargement due to hematological conditions. This dataset includes high quality CT volumes with detailed organ masks. Many researchers use spleen datasets as part of larger abdominal segmentation pipelines involving multi-organ models. The AMOS dataset provides a comprehensive benchmark for multi-organ abdominal segmentation.
The dataset’s straightforward anatomical structure makes it ideal for validating new architectures or preprocessing pipelines. It is often combined with other abdominal datasets to support multi-label tasks, contributing to more holistic models that can handle whole-body imaging.
Kidney and Tumor Segmentation (KiTS)
The KiTS dataset provides annotated CT scans of kidneys and kidney tumors, supporting both organ-level and lesion-level tasks. Tumor appearance varies significantly across cases, making KiTS a strong dataset for oncology segmentation. The dataset includes clear guidelines on how annotations were created, improving reproducibility.
KiTS has become a reference dataset for improving architectures such as 3D U-Nets, hybrid transformers, and multiscale segmentation models. Because kidney tumors often have irregular shapes and contrast differences, the dataset is useful for validating whether a model can capture complex boundaries without over-segmenting or leaking into nearby tissues.
Whole Body CT Segmentation (TotalSegmentator)
TotalSegmentator is one of the most comprehensive resources for multi-organ segmentation. It includes more than 100 segmented anatomical structures covering bones, muscles, organs, vessels, and soft tissue. This dataset is ideal for applications in biomedical research, radiotherapy planning, and anatomical modeling.
The dataset enables development of holistic models that can infer relationships between structures, improving downstream tasks like landmark detection or body composition analysis. TotalSegmentator also includes strong preprocessing pipelines and community-supported implementations that make it easy to integrate into research workflows.
Breast Ultrasound Lesion Segmentation
Breast ultrasound datasets offer unique segmentation challenges due to speckle noise, heterogeneous tissue patterns, and irregular tumor margins. Public datasets include lesion masks for both benign and malignant findings, providing valuable training material for AI systems aimed at early breast cancer detection. Ultrasound segmentation helps models distinguish between subtle edge transitions and shadow artifacts.
Because breast ultrasound is operator dependent, datasets often include variability in probe positioning and acquisition technique. This variability improves the generalizability of segmentation models deployed in real clinical environments.
Dental and Maxillofacial Segmentation Datasets
Dental CBCT datasets provide segmentation labels for teeth, mandible, maxilla, and anatomical landmarks. These datasets support orthodontic planning, implant assessment, and surgical simulation. Dental segmentation tasks often require extremely precise boundaries because millimeter-level errors can alter treatment decisions.
CBCT imaging characteristics differ markedly from CT, requiring models to adapt to different voxel intensities and noise patterns. Detailed segmentation datasets help researchers explore architectures that perform well on lower radiation imaging modalities.
Histopathology and Cell Segmentation Datasets
Digital pathology datasets support segmentation at the microscopic level, which differs significantly from radiology tasks. These datasets often include nuclei, glands, mitotic figures, and cellular boundaries, enabling detailed cell counting and phenotyping. Many digital pathology datasets use high-resolution whole slide images with gigapixel dimensions.
Multi-Organ Nuclei Segmentation (MoNuSeg)
MoNuSeg is a widely used cell segmentation dataset that focuses on nuclei boundaries across multiple tissue types. It includes diverse staining variations and imaging conditions, making it ideal for evaluating stain normalization and color augmentation strategies. Models trained on MoNuSeg often contribute to downstream classification and cell quantification tasks. Projects involving histopathology segmentation can also reference the Camelyon16 challenge, which provides high resolution whole slide images with finely annotated metastasis regions.
TNBC Mitotic Figures
Datasets focused on triple negative breast cancer provide labeled mitotic figures, which are critical for prognostic assessments. These datasets are small but extremely valuable for validating high magnification segmentation models that must detect rare events.
Lizard Dataset
The Lizard dataset includes multi-class glandular structures and nuclei labels, supporting multi-label segmentation tasks that reflect the complexity of tissue architecture. It provides challenging histology images with diverse staining styles and detailed annotations. Additional cellular and microscopy datasets can be found through the Allen Brain Atlas, which offers imaging resources for neuron and nuclei segmentation.
Ophthalmology Segmentation Datasets
Ophthalmology datasets play an important role in AI research due to the structured appearance of retinal layers and lesions. These datasets often include multi-modal imaging such as OCT, fundus photography, and fluorescein angiography.
Retinal OCT Layer Segmentation
OCT datasets include pixel-level masks of retinal layers that are used for diagnosing macular degeneration, diabetic retinopathy, and glaucoma. OCT segmentation requires precise boundary detection and is highly sensitive to noise.
Fundus Lesion Segmentation
Fundus datasets include masks for microaneurysms, hemorrhages, and neovascularization. These datasets help support early screening systems for diabetic retinopathy and other vascular diseases.
Colon Polyp Segmentation Datasets
Endoscopy datasets focused on colon polyp segmentation support early detection of colorectal cancer. These datasets contain frames from colonoscopy videos, together with pixel-level segmentation masks for polyps. They pose unique challenges related to lighting variability, motion blur, and mucosal reflection.
Many research teams use these datasets to evaluate models designed for real-time inference during endoscopic procedures. These datasets contribute to the development of clinical decision support systems aimed at improving polyp detection rates.
Lung Nodule and Airway Segmentation Datasets
CT datasets focused on lung anatomy provide masks for airways, nodules, lobes, and fissures. They support research on lung cancer screening, pulmonary function analysis, and chronic disease monitoring.
Models trained on these datasets often face challenges related to small object segmentation, especially when nodules are only a few millimeters in size. Accurate segmentation helps improve downstream detection systems and supports quantitative imaging biomarkers.
Choosing the Right Dataset for Your Clinical AI Project
Selecting the right medical image dataset depends entirely on the clinical goal, modality, and expected deployment conditions. Developers must consider imaging modality, anatomical region, pathology type, availability of metadata, and the quality of the ground truth.
Match the Dataset to the Imaging Modality
Projects involving MRI, CT, ultrasound, or digital pathology each require datasets with appropriate acquisition characteristics. Using an MRI dataset to train a CT-based model is rarely effective because intensity distributions and anatomical appearance differ markedly. Teams should ensure that dataset modalities match real clinical workflows.
Consider Annotation Quality and Clinical Review
A well structured medical image dataset should include clear documentation of annotation protocols. When expert consensus or multi-annotator review is used, labels are more likely to reflect clinically meaningful boundaries. Researchers should review dataset documentation carefully to understand its limitations.
Evaluate Dataset Size and Diversity
Large datasets provide better statistical coverage of clinical variation. However, small high-quality datasets created with expert involvement can still be extremely valuable. Teams often combine multiple datasets or use transfer learning to improve model performance.
Assess Preprocessing Requirements
Datasets with strong documentation reduce preprocessing complexity and improve reproducibility. Teams should consider whether the dataset includes voxel spacing, normalization guidelines, or suggested splits.
Balance Public and Private Data
While public datasets are excellent for benchmarking, clinical deployments often require private, institution-specific data for fine-tuning. Combining both types can significantly improve generalization and reduce bias.
The Role of Large Multi-Institutional Datasets in Advancing AI
As medical imaging evolves, multi-institutional datasets play an increasingly important role in building reliable AI systems. Datasets that include multiple hospitals, imaging vendors, and diverse patient demographics offer robust training foundations. They help mitigate issues related to clinical bias, scanner differences, and population variability.
Large scale datasets also enable training of foundation models in medical imaging. These models learn generalizable representations that can be adapted to multiple segmentation tasks, reducing the need for large annotated datasets for each application. This trend is accelerating the development of clinically useful models across radiology, oncology, and digital pathology.
Emerging Trends in Medical Segmentation Datasets
The next generation of datasets will likely integrate multimodal inputs, weak labels, synthetic data, and self-supervised representations.
Multimodal Imaging
Datasets combining CT, MRI, PET, and histology are becoming more common. These datasets allow cross modality learning, where features learned in one modality improve performance in another.
Weak and Semi-Supervised Labels
Manually annotating large volumes of medical data is resource intensive. Weak supervision, coarse labels, and self-training pipelines help scale dataset size without sacrificing too much clinical accuracy.
Synthetic Training Data
Synthetic data generated using generative models or physics based simulations supports rare pathology modeling and helps balance datasets with class imbalance.
Federated Datasets
Federated datasets enable training across multiple hospitals without centralizing patient data. This approach addresses privacy concerns and encourages collaboration.
If You Are Working on Medical Imaging Segmentation
If you are planning an AI project involving segmentation, careful dataset selection is critical. Teams should evaluate task complexity, clinical requirements, annotation quality, and available benchmarks. Combining strong public datasets with high quality private data often produces the most reliable results.
If you are working on an AI or medical imaging project, our team at DataVLab would be glad to support you.